Bioinformatics Pipeline
Bioinformatics pipeline that automates the processing of large biological datasets.
🧪 Overview
This example a bioinformatics pipeline depicts a series of orchestrated data processing steps where each step builds on the previous one. These pipelines are crucial in dealing with the influx of large biological data, particularly in genomics, proteomics, or complex interactions with them. They offer reproducibility, organization, and automation, enabling the processing of large-scale biological data and achieving reproducible results.
✏️ Steps in a Typical Bioinformatics Pipeline:
- Data Collection: Collect raw data from genomic sequencing, usually from a sequencing machine, which consists of short DNA sequences called reads.
- Data Preprocessing: Process raw data to ensure the quality of reads, correct or remove low-quality reads, and prepare them for downstream analysis such as read mapping.
- Data Analysis: Perform the main analysis tasks such as variant calling, identifying genetic variations within the sequenced genomes.
- Interpretation and Visualization: Interpret the results of the analysis, annotate variants to provide information about their potential effects, and visualize the results using plots or tables.
Building Bioinformatics Pipelines
There are several ways to build bioinformatics pipelines:
- Scripting Languages: Use scripting languages like Python or Bash to write a series of scripts that perform each step of the pipeline.
- Workflow Management Systems: Utilize workflow management systems such as
Snakemaketo define a series of rules that describe how to create the final output from the input data. - Automated GUI: Use automated graphical user interfaces like Galaxy to construct pipelines by connecting predefined tools and workflows.
🐍 Snakemake Pipeline for DNA Sequence Mapping
This Snakemake pipeline automates DNA sequence mapping using the BWA aligner, facilitating the alignment of sequencing reads to the human reference genome (hg19_chr8.fa) for multiple samples (father, mother, and proband). The pipeline uses the parallel processing for efficient handling of large-scale sequencing data.
📂Project Structure
The project directory is organized as follows:
📂snakemake_pipeline/
├── 📂fastq/
│ ├── father_R1.fq.gz
│ ├── father_R2.fq.gz
│ ├── mother_R1.fq.gz
│ ├── mother_R2.fq.gz
│ ├── proband_R1.fq.gz
│ └── proband_R2.fq.gz
├── 📂ref/
│ └── hg19_chr8.fa
├── links.txt
├── 📂mapped_reads/
├── 🐍Snakefile
├── 🐍Snakefile2
└── 🐍Snakefile3
- links.txt: A file containing links to FASTQ files containing DNA sequencing reads and the genome reference.
- 📂ref/: Directory containing the reference genome file (
hg19_chr8.fa). - 📂fastq/: Directory containing the raw FASTQ files for sequencing data.
- 📂mapped_reads/: Directory for storing the mapped sequencing reads in BAM format.
- 🐍Snakefile, Snakefile2, Snakefile3: Snakemake workflow files for different stages of the pipeline.
🎯 Snakemake Workflow Explanation
🐍 Snakefile
The Snakefile defines a rule named bwa_map that maps sequencing reads from the father’s FASTQ files to the reference genome using BWA. The output is stored as mapped_reads/father.bam. It utilizes fixed file paths for the reference genome and father’s FASTQ files.
🐍 Snakefile2
Similarly, Snakefile2 defines a rule named bwa_map to map sequencing reads from any sample’s FASTQ files (specified by {sample}) to the reference genome. The output is stored as mapped_reads/{sample}.bam. This rule allows for mapping multiple samples in a flexible manner.
Additionally, there’s a cleanup rule to remove all BAM files from the mapped_reads/ directory after completion.
🐍 Snakefile3
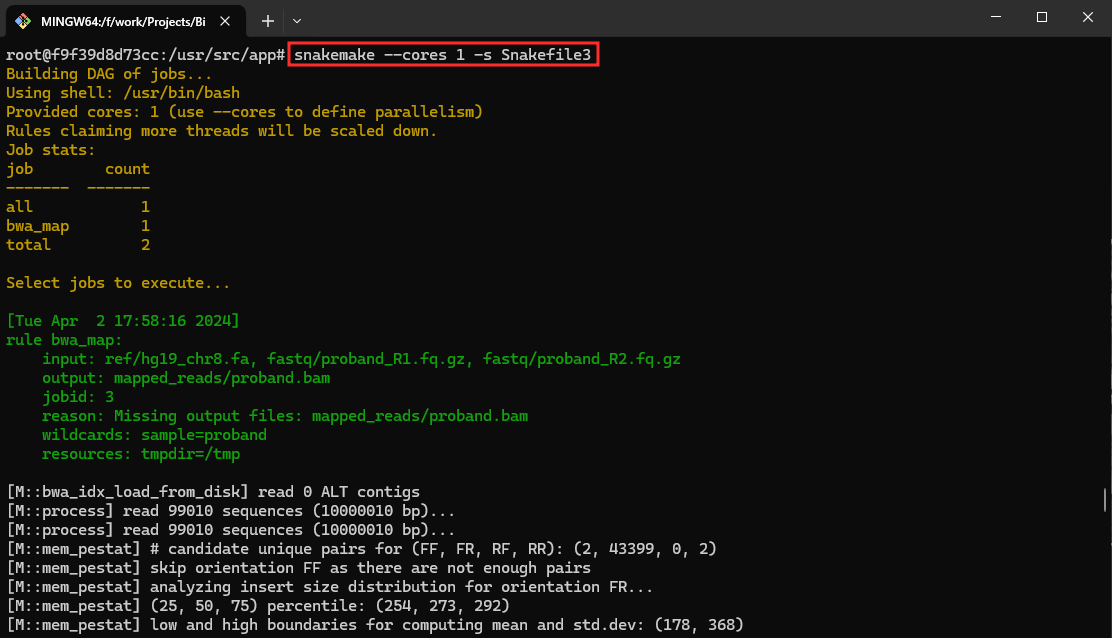
Snakefile3 defines a list of samples [father, mother, proband]. The rule all ensures that all samples are processed, and the bwa_map rule is applied to each sample’s FASTQ files. Similar to Snakefile2, it also includes a cleanup rule.
📝 Interpreting Results
After executing the Snakemake workflow, the mapped sequencing reads will be stored in BAM format in the mapped_reads/ directory. These BAM files can be further analyzed using tools like Samtools or visualized using genome browsers to interpret the alignment results.
🚀 Installation
- Ensure
BWAandSamtoolsare installed in your environment.
sudo apt-get install bwa
sudo apt-get install samtools
conda activate snakemake
# Fetch data using links provided
cat links.txt | xargs -i -P 4 wget '{}'
- Place the reference genome file (
hg19_chr8.fa) in theref/directory. - Place the FASTQ files for each sample in the
fastq/directory. - Customize the Snakefile (
Snakefile), Snakefile2 (Snakefile2), or Snakefile3 (Snakefile3) as per your requirements. - Execute Snakemake using the desired Snakefile:
snakemake -s Snakefile.
🛠️ Usage
Pipeline Execution with Snakemake
Below are the steps involved in executing a pipeline using Snakemake:
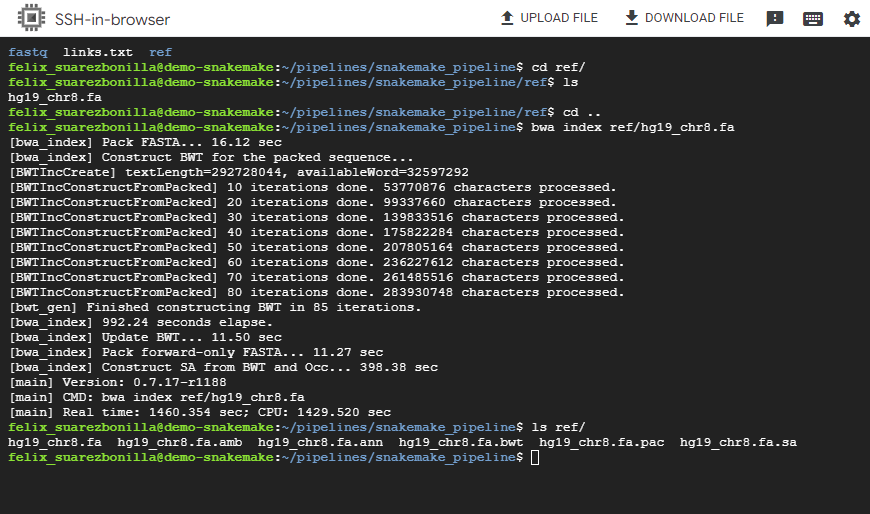
- Index the reference genome using BWA.
bwa index ref/hg19_chr8.fa
This command indexes the reference genome file hg19_chr8.fa using the BWA aligner. Indexing is a necessary step for efficient alignment of sequencing reads.
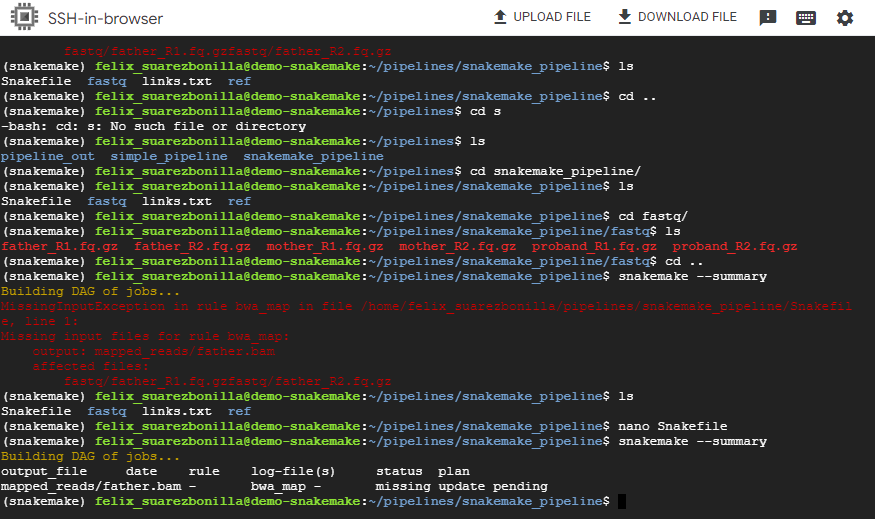
- Show the snakemake summary
snakemake --summary
This command generates a summary of the Snakemake workflow, showing information such as the number of rules, targets and files to be created.
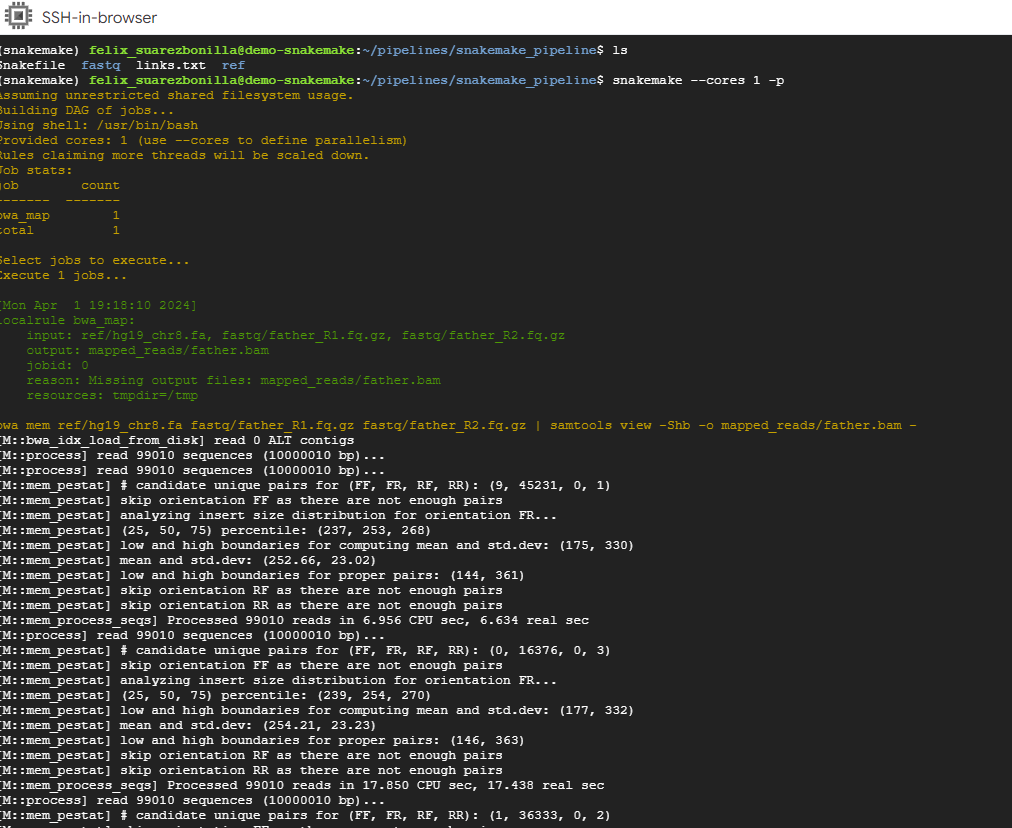
- Execute the pipeline using Snakemake.
snakemake --cores 1 -p
This command executes the Snakemake workflow using a single core (–cores 1) and prints the commands that are executed (-p).
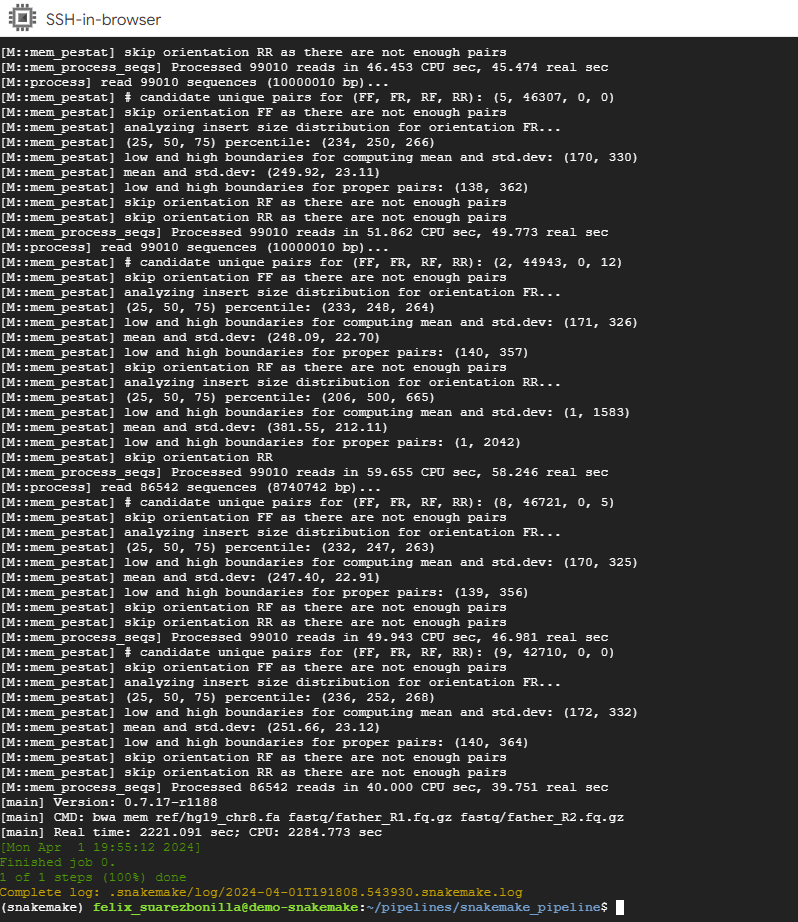
- Monitor and manage pipeline execution.
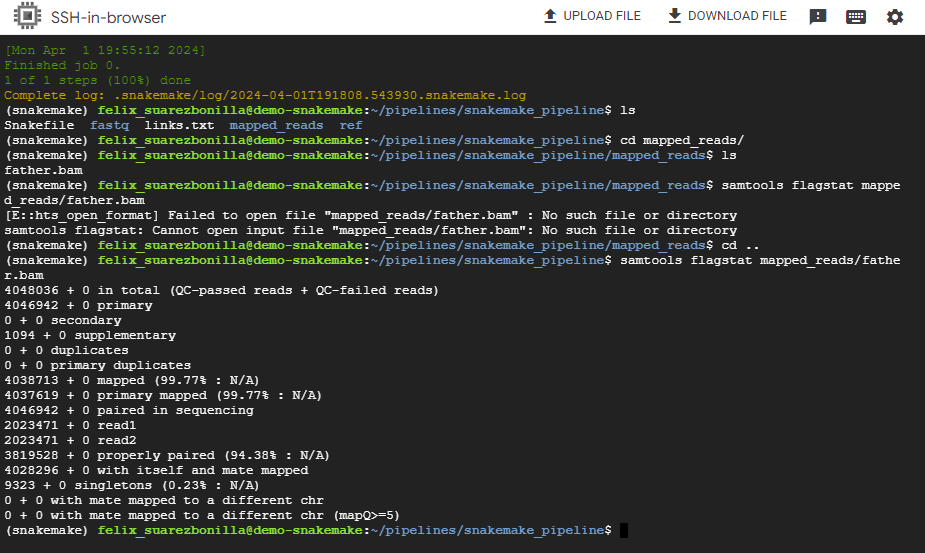
samtools flagstat mapped_reads/father.bam
This command uses samtools to generate flag statistics for the BAM file father.bam, which likely contains alignment information for sequencing reads from a sample named “father”.
Pipeline Execution with Snakemake2
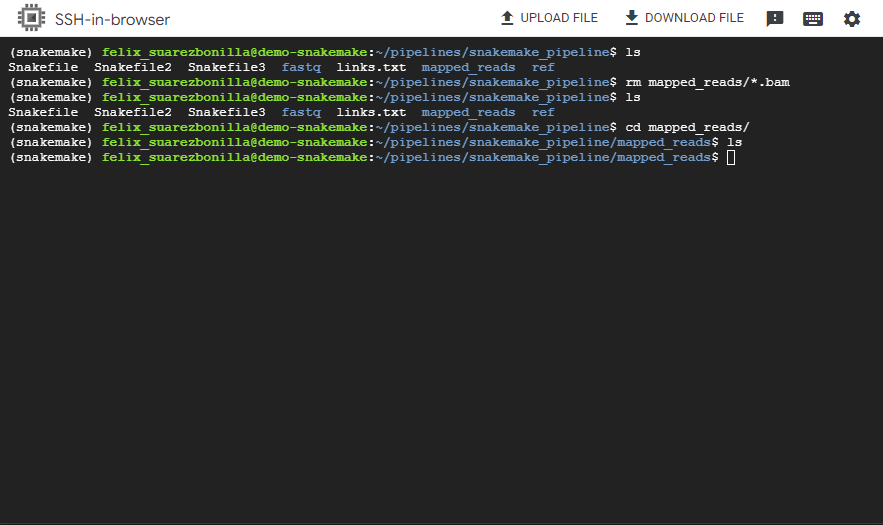
- Remove the mapped_reads folder
rm mapped_reads/*.bam
This command removes all BAM files from the mapped_reads/ directory.
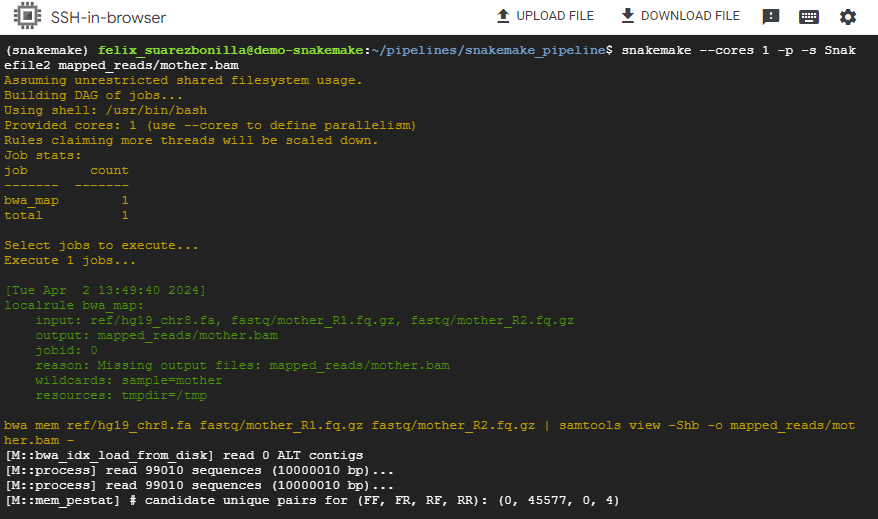
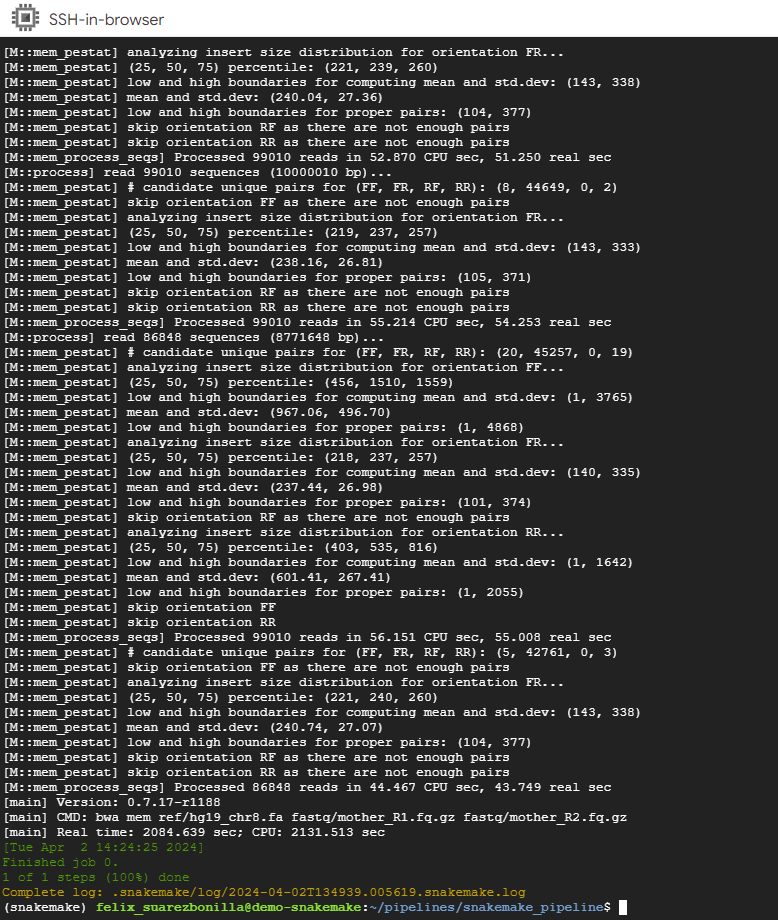
- Execute the pipeline Snakefile2
snakemake --cores 1 -p -s Snakefile2 mapped_reads/mother.bam
This command executes the Snakemake workflow using a single core and focuses on generating the output file mother.bam.
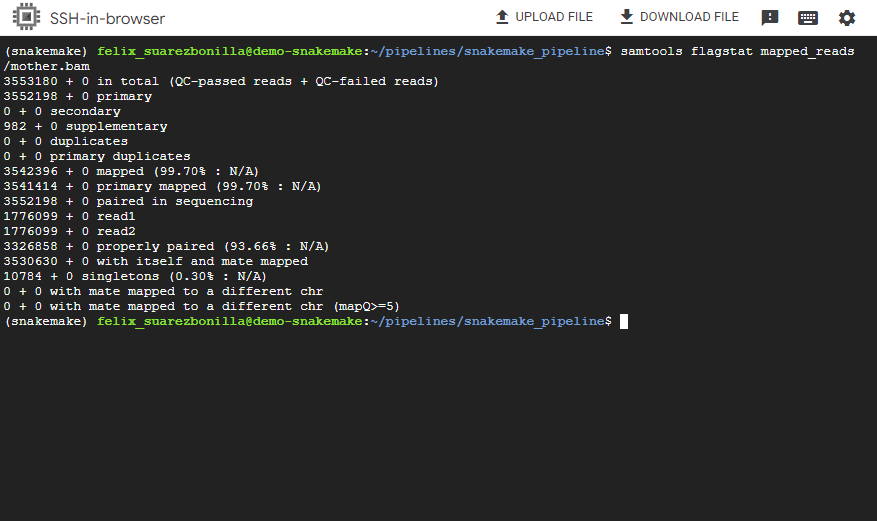
- Visualization of retuls with samtools
samtools flagstat mapped_reads/mother.bam
This command the samtools tool to compute flag statistics for the BAM file named mother.bam. This file stores alignment data pertaining to sequencing reads originating from a biological sample labeled as mother.bam
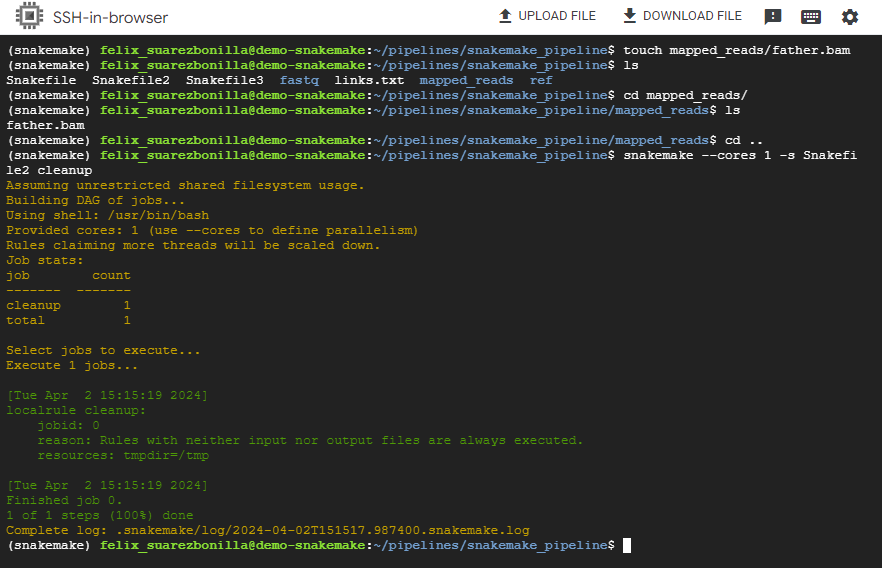
- Perform cleanup operations
snakemake --cores 1 -s Snakefile2 cleanup
This command executes the Snakemake workflow specified in Snakefile2 and performs cleanup operations.
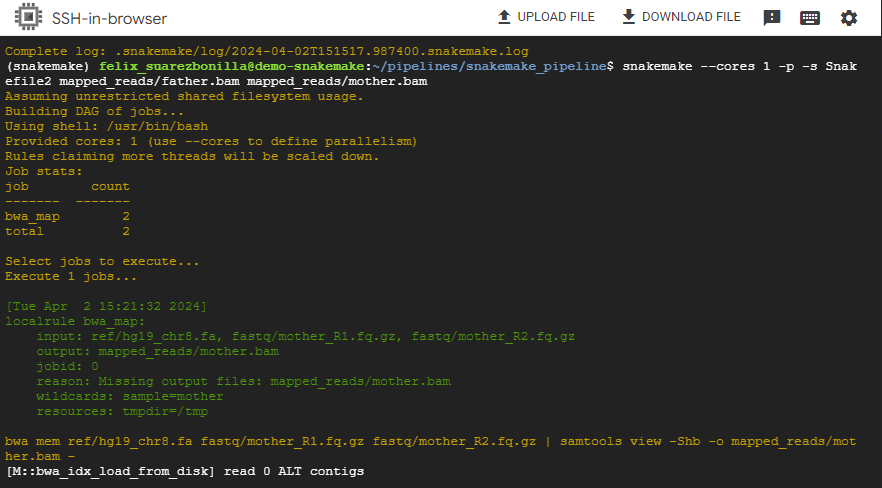
- Generate multiple outputfiles
snakemake --cores 1 -p -s Snakefile2 mapped_reads/father.bam mapped_reads/mother.bam
This command executes the Snakemake workflow specified in Snakefile2 and focuses on generating the output files father.bam and mother.bam.
Pipeline Execution with Snakemake3
- Remove the mapped_reads folder
snakemake --cores 1 -s Snakefile2 cleanup
This command executes the Snakemake workflow specified in Snakefile2 and performs cleanup operations.

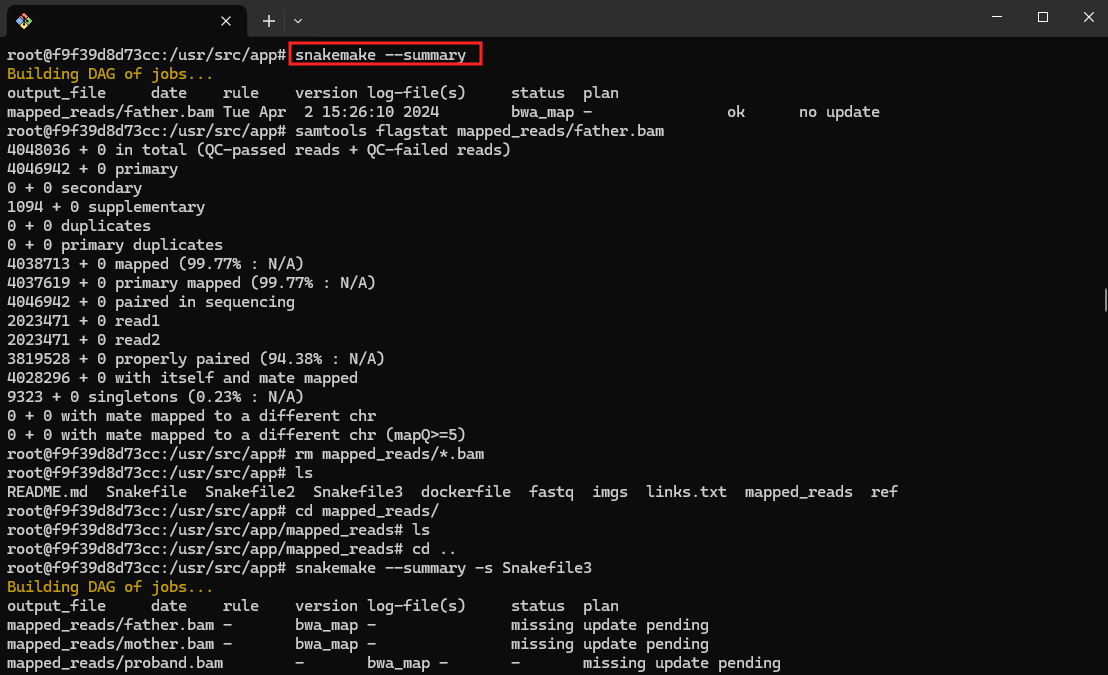
- Show the snakemake summary
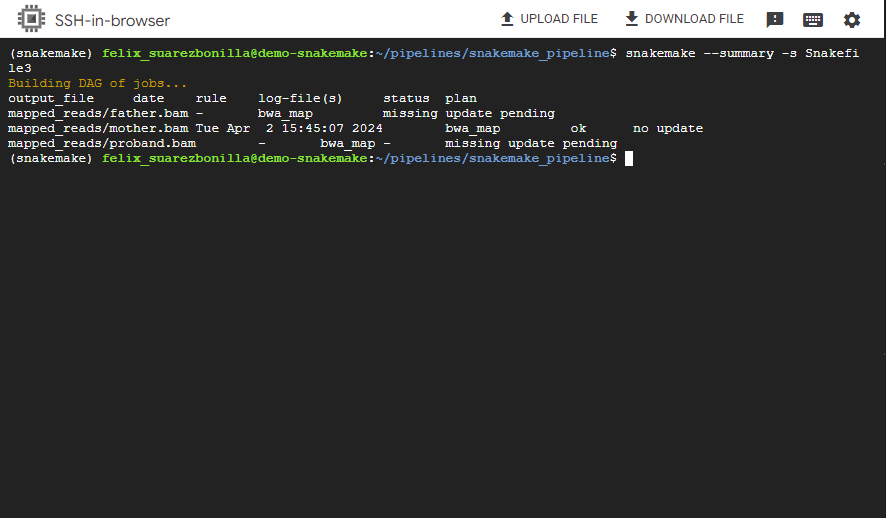
snakemake --summary -s Snakefile3
This command generates a summary of the Snakemake workflow, showing information such as the number of rules, targets and files to be created.
- Execute the pipeline Snakefile3
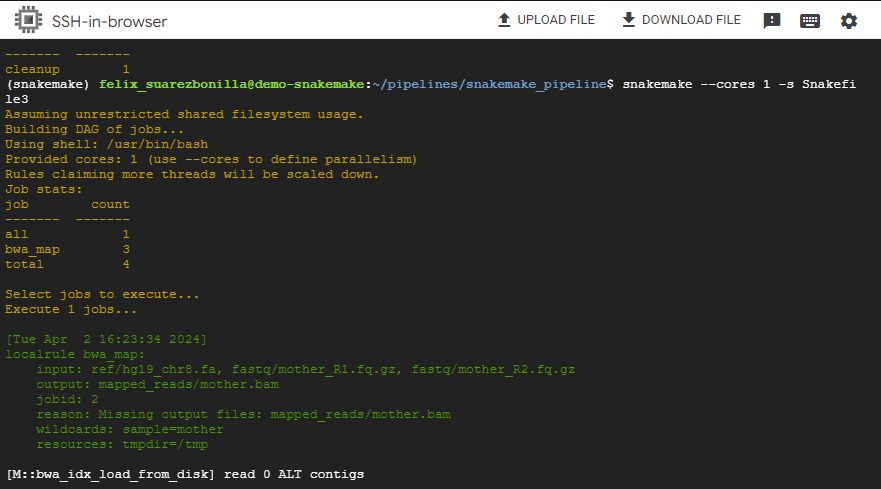
snakemake --cores 1 -s Snakefile3
This command executes the Snakemake workflow using a single core and focuses on generating the output files father.bam, mother.bam and proband.bam.
🐳 Dockerimage
To containerize the bioinformatics pipeline, Docker can be utilized. Below are the steps to build and run the Docker image:
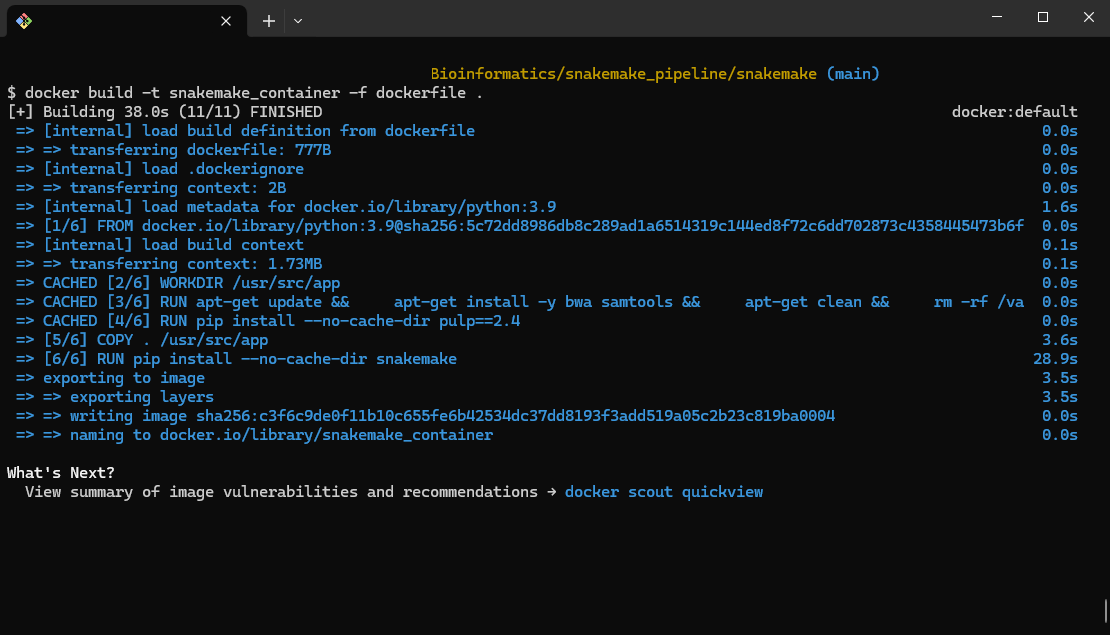
- Building the Docker Image:
docker build -t snakemake_container -f dockerfile .
This command builds a Docker image named snakemake_container using the provided dockerfile.
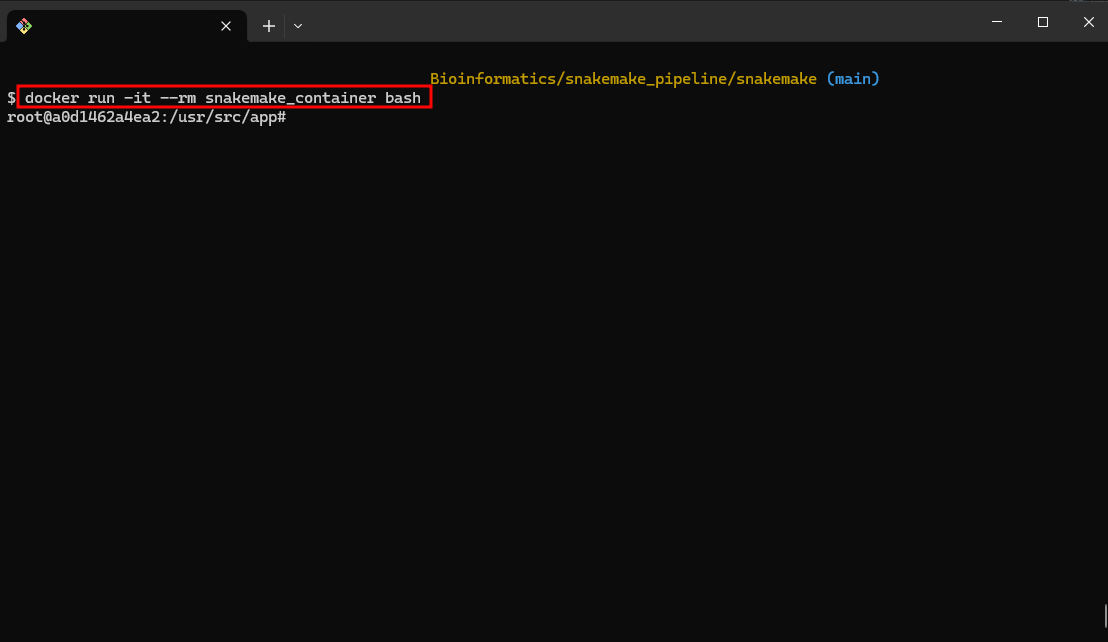
- Run the Docker Container:
docker run -it --rm snakemake_container bash
This command runs a Docker container interactively using the snakemake_container image, providing a Bash shell.
- Index the Reference Genome:
bwa index ref/hg19_chr8.fa
Index the reference genome file hg19_chr8.fa using the BWA aligner.
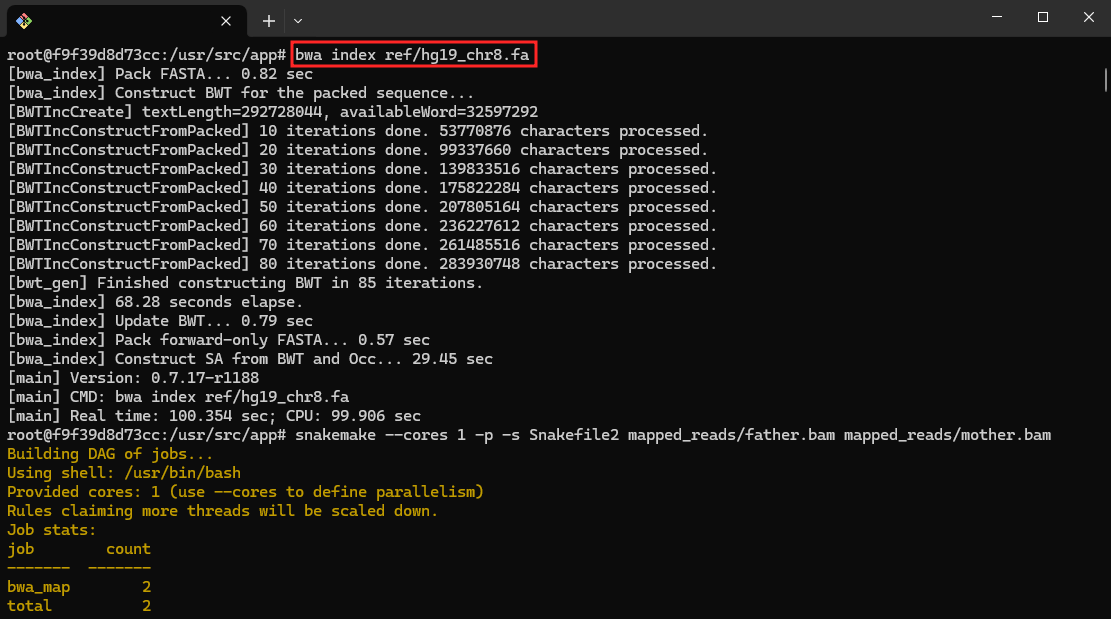
- Generate Snakemake Summary:
snakemake --summary
This command generates a summary of the Snakemake workflow.
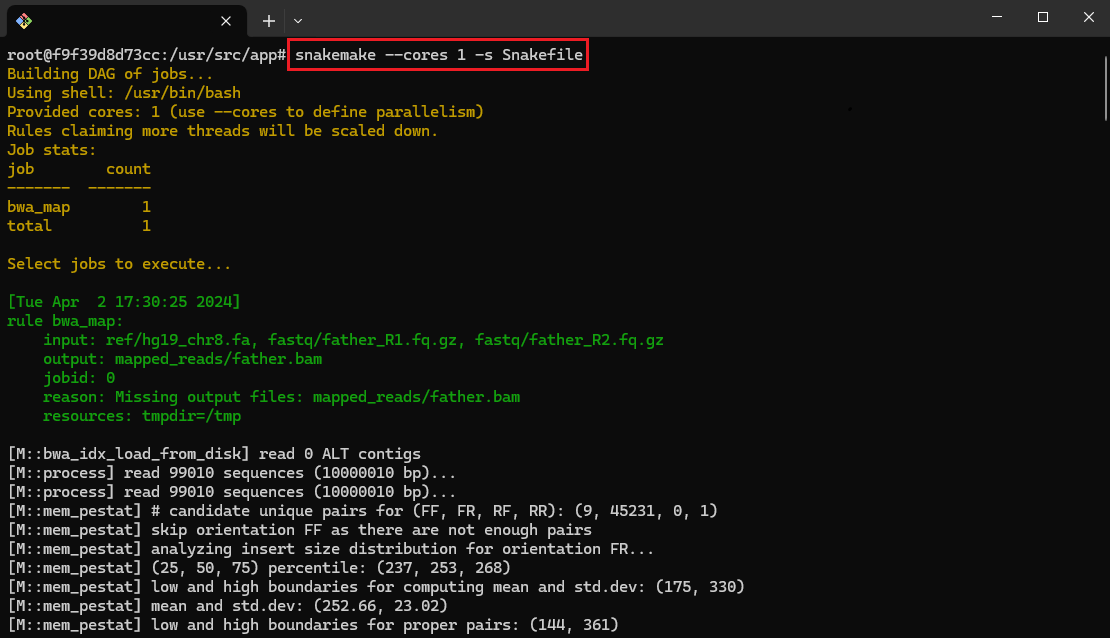
- Execute Snakemake Workflow:
snakemake --cores 1 -s Snakefile
This command executes the Snakemake workflow using a single core.
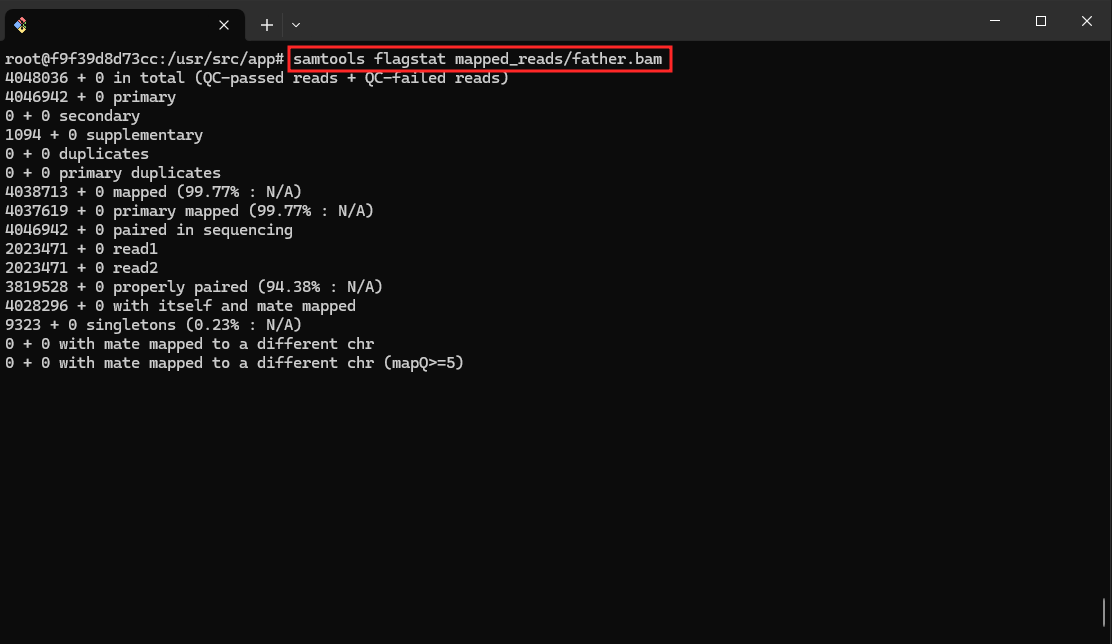
- Visualize Results with Samtools:
samtools flagstat mapped_reads/father.bam
This command generates flag statistics for the BAM file father.bam.
- Remove mapped_reads:
rm mapped_reads/*.bam
This command removes all BAM files from the mapped_reads/ directory.
- Execute Snakefile2 Workflow:
snakemake --cores 1 -p -s Snakefile2 mapped_reads/mother.bam
This command executes Snakefile2, focusing on generating the output file mother.bam.
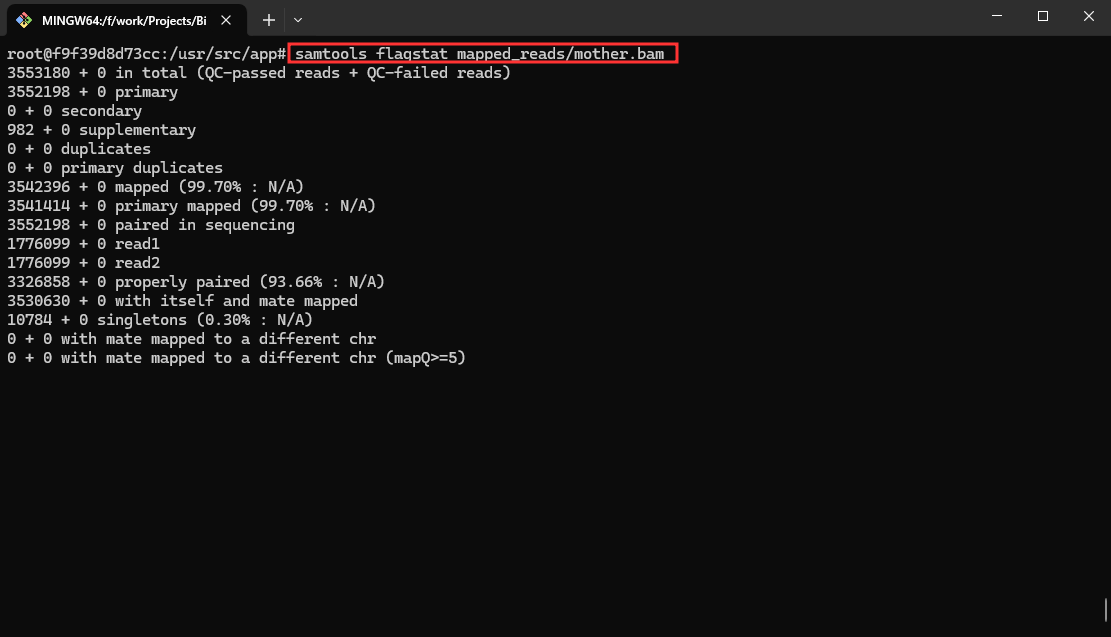
- Visualize Results with Samtools (Snakefile2):
samtools flagstat mapped_reads/mother.bam
This command generates flag statistics for the BAM file mother.bam.
- Cleanup Operation (Snakefile2):
snakemake --cores 1 -s Snakefile2 cleanup
This command performs cleanup operations specified in Snakefile2.
- Execute Multiple Outputs (Snakefile2):
snakemake --cores 1 -p -s Snakefile2 mapped_reads/father.bam mapped_reads/mother.bam
This command executes Snakefile2, focusing on generating the output files father.bam and mother.bam.
- Execute Snakemake Workflow (Snakefile3):
snakemake --cores 1 -s Snakefile3
This command executes the Snakemake workflow defined in Snakefile3.